An MIT team is getting to know a little-studied bacterium that consumes materials ranging from soil contaminants to corn husks and stores them as globules of fat ideal for making biofuels. Using a variety of approaches, the researchers have been gaining insights into the biological processes and genetic pathways that control that conversion, enabling them to further enhance the bacterium’s fat-making ability.
Best of all, they are converting the fat into fuels that—unlike ethanol—contain enough energy per liter to be used in trucks, trains, ships, and even airplanes. “We’re already making fat from our bacteria into jet fuel,” says Anthony Sinskey, professor of biology and director of the research. “We’re going to fly an airplane on it!”
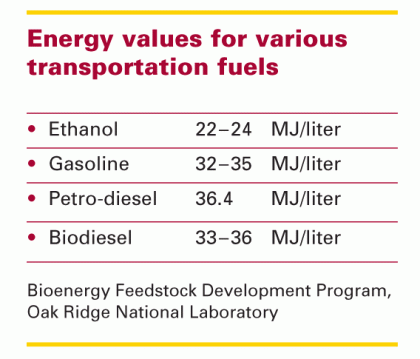
As the nation struggles to replace fossil fuels used in transportation, much attention has focused on ethanol and hydrogen. But Sinskey’s team is instead aiming for biodiesel, a fuel with several advantages. It has a much higher energy density than ethanol has (see the chart at the right). It does not have the national security issues—or the pollutant emissions—associated with diesel made from petroleum. And unlike hydrogen, it is a “drop-in” fuel. “We already have the infrastructure for transporting, storing, and pumping biodiesel and the vehicle engines that can burn it,” says Sinskey.
Most people making biodiesel start with a fat called triacylglycerol, or TAG. Familiar sources of TAGs include leftovers from deep fat fryers and grease traps, fats from meat rendering, and vegetable oils. The conversion to biodiesel is so simple that people are doing it in their garages and barns, according to Daniel MacEachran, a postdoctoral associate in the Department of Biology. As a bonus, TAGs can also be converted into jet fuel. “But we’re never going to power the country or the world on grease traps and animal renderings,” says MacEachran. Growing algae on biomass is another non-food approach to producing TAGs, but the technology is still under development, and there are problems to be solved.
Fat-producing bacteria
Another natural source of TAGs is bacteria. Give bacteria lots of carbon (crop waste and the like) and not much nitrogen, and they will store excess carbon for future use. In about 90% of bacteria, the stored carbon is in the form of a polymer, which some members of the Sinskey Lab are using to make “bioplastics.”
But a small group of bacteria instead stores the carbon in the form of TAGs, and one of them that does it really well is Rhodococcus opacus. This bacterium is not well understood and is more difficult to work with than, say, E. coli. But it grows quickly; it is not pathogenic; and it produces remarkable quantities of TAGs—in some tests, as much as 75% of its dry weight. Moreover, it can metabolize a wide range of carbon sources. Indeed, it was first found growing on hydrocarbon contaminants in sand at a gas works facility in Germany. “So these bacteria not only survive but actually thrive on compounds that would kill most other organisms,” says MacEachran.
Expanding their appetites
In the ideal fermentation process, Rhodococcus would quickly and completely consume mixtures of carbon “feedstocks” and produce large quantities of TAGs. But there are some things that even this bacterium does not eat. One of them is xylose, a key constituent—along with glucose—of the cell walls of plants. If Rhodococcus bacteria are going to consume cellulosic sources such as corn stover and switchgrass, they need to metabolize xylose as well as glucose.
To achieve that goal, Kazuhiko Kurosawa, a research scientist in biology, turned to genetic engineering. He took carefully chosen genes from bacteria that naturally metabolize xylose and inserted them into Rhodococcus. The result: a strain of Rhodococcus that metabolizes xylose to produce TAGs. Better still, this new strain consumes glucose and xylose at the same time and at about the same rate. Most bacteria, if presented with two carbon sources, will consume first one and then the other, with a lag in growth as they switch gears in the middle. Because Kurosawa’s organism “co-utilizes” xylose and glucose, there is no pause to switch gears, so the overall rates of growth and fat production are dramatically higher—and all the feedstock is consumed.
In addition, the bacterium makes about the same amount of TAGs on the mixture as it does on glucose or xylose alone, and the composition of the TAGs is the same. “So there must be some common intermediate that’s formed from all carbon feedstocks, and that intermediate—not the feedstock itself—controls the TAGs synthesis,” says Sinskey. “That’s an interesting phenomenon.”
Identifying fat-making genes
Another approach to increasing Rhodococcus‘s ability to metabolize carbon and make fat is to work with the bacterium’s own genes. The challenge there is to determine which genes play a role in producing and storing TAGs and to define exactly what each gene or set of genes does. As a critical first step, Jason Holder, postdoctoral associate in biology, sequenced the genome of Rhodococcus opacus, identifying all of the DNA and more than 7,000 genes that make up its chromosome.
MacEachran then developed a genetic screening technique to locate the genes of interest. The technique involves the use of a “transposon,” a short stretch of DNA that can integrate anywhere among the genes on a chromosome. The presence of the transposon disrupts the function of whatever gene it happens to land in. So if a Rhodococcus bacterium is exposed to a transposon and then stops or slows its production of TAGs, the transposon has landed in a gene essential for TAG metabolism. Sequencing the DNA of that bacterium will show where the transposon—with its recognizable DNA—is located and therefore which gene has been affected.
But many genes are likely to play a role in TAG metabolism, so MacEachran developed a “high-throughput” screening technique. He uses a transposon that carries resistance to the antibiotic kanamycin. He takes a batch of bacteria—billions of individual cells—and mixes them with a batch of transposons. Most of the bacteria cells will remain unchanged, but a small fraction will pick up a transposon. When the cells are exposed to kanamycin, only those containing a transposon will be immune to the kanamycin and so will live.
In a painstaking process, the researchers then separate the remaining cells, spread them out on a plate with glucose, and let them divide and replicate to form distinct colonies. Each colony will contain tens of thousands of identical cells, every one with a transposon in the same location in its genome. The colonies are next grown under carefully optimized high-carbon, low-nitrogen conditions and then stained with a chemical dye that is readily absorbed by fat. Most of the colonies become dark—a sign that the transposon has not interfered with their production of TAGs. But a few will remain pale. In those colonies, the transposon has lodged in and disrupted a gene that plays some critical role in the formation of TAGs.
Key genes: What do they do?
That screening technique has permitted the researchers to identify a set of genes that affect TAG formation. “But now we have to do the hard science of characterizing each gene and figuring out exactly what role it plays in TAG metabolism,” says MacEachran.
He has been particularly intrigued by a gene he calls “tadA” (from “TAG accumulation defective”). Genes similar to the tadA gene have been found in several other organisms that also make fat, but its exact function is unknown. “No one else has worked on tadA before, so we’re moving into new territory,” says MacEachran.
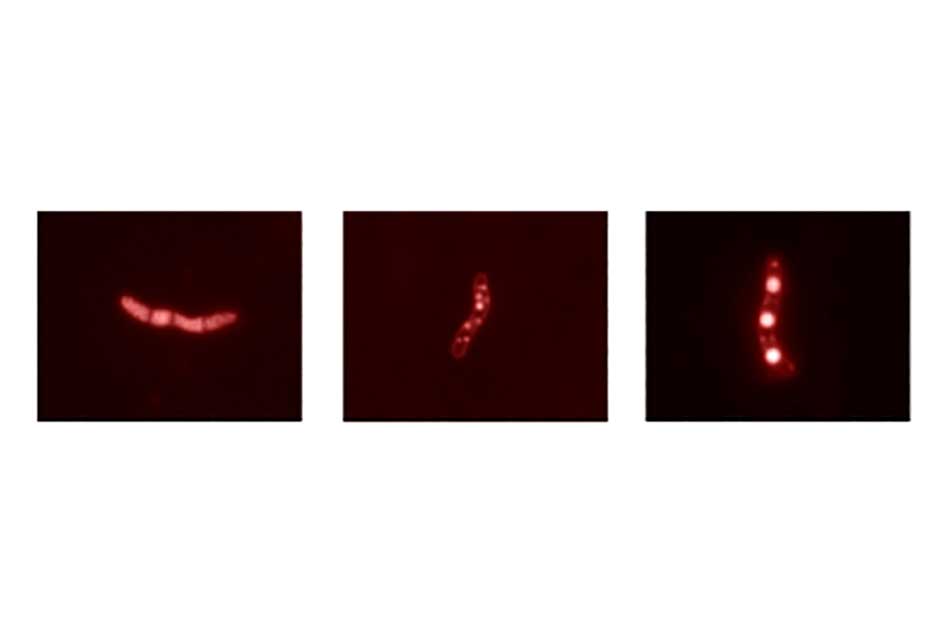
To get a closer look at tadA, he uses a technique called fluorescent microscopy combined with a dye called Nile red. Within a bacterium, TAGs are contained in large spherical structures known as lipid bodies. Those lipids readily take up Nile red and become fluorescent, thus easily differentiated from the rest of the cell under the microscope.
The images above right show three types of Rhodococcus bacteria, each with its lipid bodies appearing as glowing spheres. At the left is the “wild type,” a natural, unaltered Rhodococcus bacterium. The lipids are fairly uniform in size and distribution. The bacterium in the middle image lacks the tadA gene. Now there are both small and large lipids, and rather than being uniformly distributed, they appear stuck to the walls. The right-hand image shows a bacterium that contains lots of extra tadA gene. Now there are massive lipid bodies that sit right down the middle of the cell.
Taken together, these observations suggest that the tadA gene’s primary role may not be in making fat but rather in storing it. “Losing the tadA gene seems to mess up the organism’s ability to assemble the lipids properly,” says MacEachran. “And having too much of it produces big, single lipid bodies. So we believe that this gene produces a protein that acts as sort of a shepherd to pull all of the lipid bodies together into fewer, larger bodies.”
Continuing investigations
The researchers are continuing to gain insights into the workings of their bacterium. Studies of “neighbors” of tadA show that those genes also influence lipid body structure, each one in a slightly different way. And analyses of similar genes in other organisms suggest a model for certain Rhodococcus genes that may help lipid bodies attach to one another and coalesce. Further screening has identified genes involved in the actual production of TAGs.
The feedstock studies also continue. The researchers have now tested almost 200 carbon sources, looking for low-cost, high-yield ones that could improve process economics. They have identified specific components in feedstocks that inhibit cell growth and interfere with TAG production. And they have engineered a strain of Rhodococcus that metabolizes glycerol, a waste product of biodiesel production. With the engineered strain, the glycerol could be recycled as a feedstock for more biodiesel production.
Finally, Sinskey and his team are working with industry to develop TAG-extraction methods that will yield highly enriched TAG fractions and low concentrations of products known to interfere with the conversion of TAGs into biodiesel. In addition, they are examining chemical and biological catalysts for converting TAGs to fuels and are working to scale up production.
If all goes as planned, Sinskey estimates that within a few years the Rhodococcus-based biofuels will be commercially available as a viable, sustainable alternative to today’s petroleum-based transportation fuels.
This research was funded by a seed grant from the MIT Energy Initiative; by Shell International Exploration & Production, Inc.; and by Logos Technologies, Inc. More information can be found in:
K. Kurosawa, P. Boccazzi, N. de Almeida, and A. Sinskey. “High glucose cultivation of Rhodococcus opacus PD630 in batch-culture for biodiesel production,” Journal of Biotechnology, in press 2010.
This article appears in the Spring 2010 issue of Energy Futures.
Press inquiries: miteimedia@mit.edu